TRANSONCO: an ONCOlille transdiciplinary projet.
The TransOnco project draws on knowledge from biology, microelectronics (BioMEMS), computer science, data science, electronics, mathematics, and physics, synergizing from several ONCOLille institut laboratories (Canther, Phycell, LPP, and LIMMS), and collaborating units from University of Lille (PhLAM, IEMN, PRISM).
Abstract of the project:
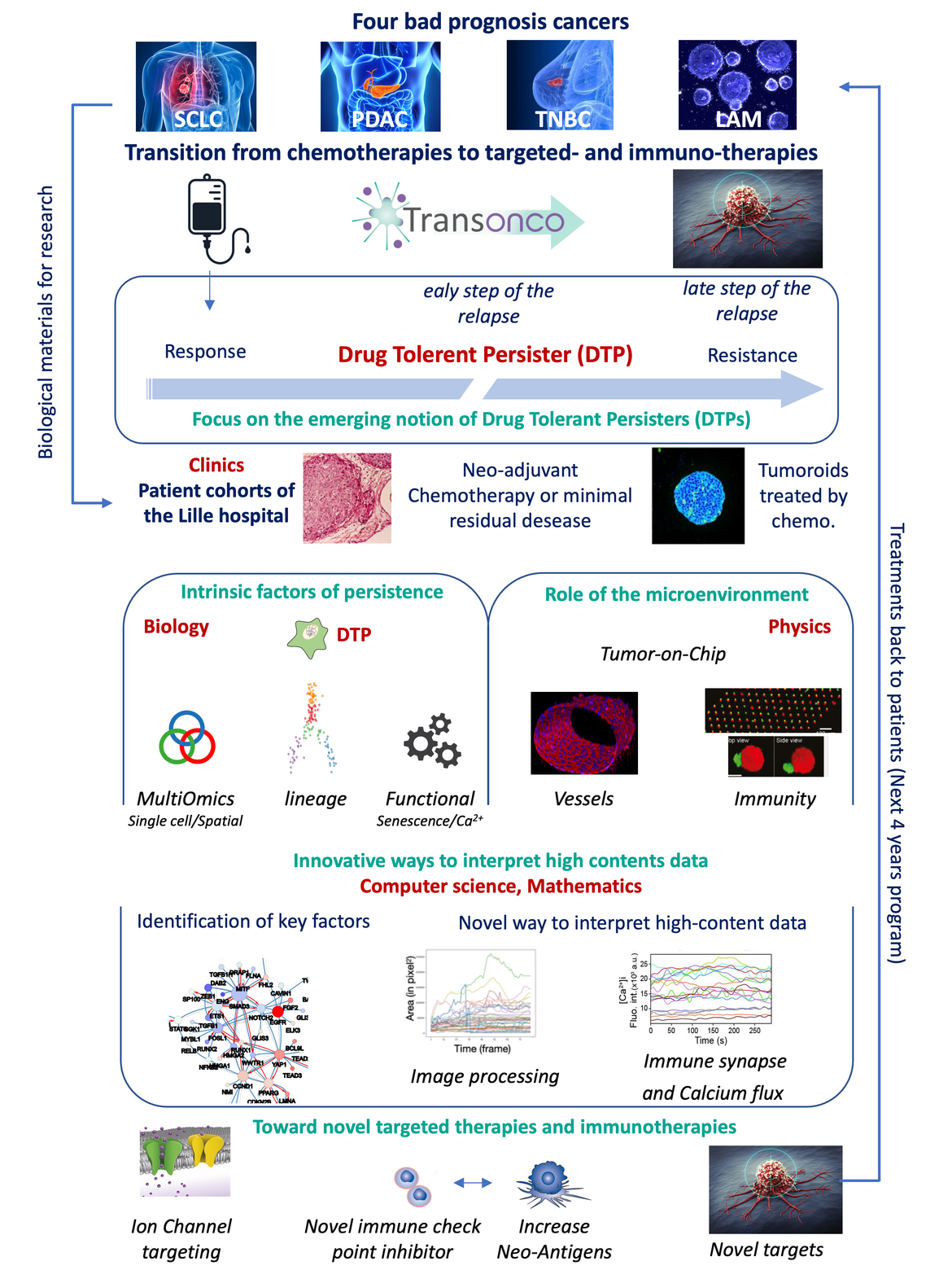
Therapies against advanced forms of cancer have seen a leap in effectiveness in the last decade, thanks in particular to the emergence of targeted therapies and immunotherapies, enabling a transition towards what is known as precision medicine. The benefit for patients materializes in an increase in life expectancy with cancer and an improvement in quality of life. However, many patients and some cancer types do not benefit from these novel therapies and are thus still treated by cytotoxic chemotherapies. Due mainly to the lack of effective treatment, which for patients translates into inevitable relapse, most of these cancers are classified as bad prognosis cancers. The TransOnco program aims to propose a transition from chemotherapies to targeted therapies and immunotherapies for four bad prognosis cancers (i) small-cell lung cancer, (ii) triple-negative breast cancer, (iii) pancreatic adenocarcinoma and (iv) acute myeloid leukemia still treated by chemotherapies. For these cancers, targeted therapies are not used or have been inoperative since effective targets were not identified, and microenvironment conditions make immunotherapies inefficient. To identify new therapeutic targets for these cancers, we propose to focus our attention on the early stages of the chemotherapeutic treatment, which display initial efficacy but lead to the establishment of a minimal residual disease (MRD) at the origin of systematic relapse. This MRD is constituted by a contingent of recently described Drug Tolerant Persisters (DTPs). This novel concept involves an immediate cellular adaptation based on a cellular plasticity that is still poorly understood and whose biological foundations remain to be deciphered.
Our project will be based on (i) the crossdisciplinary organization of the ONCOLille Institute, bringing together clinicians, biologists, physicists, and mathematicians from five Lille-based research units dedicated to the fight against cancer, (ii) the state-of-the-art technical platforms within the ONCOLille Institute and the university, and (iii) our direct interaction with Lille hospitals and access to patient cohorts with longitudinal follow-up. Since MRD is difficult to collect on patients, our aim is first to set up innovative models to reconstitute the DTPs. Established cancer cell lines and freshly tumor-derived cancer cells from our local cohorts will be cultured in 3D to derive tumoroids recapitulating tumor heterogeneity. These tumoroids will be treated by chemotherapies to produce DTPs. Clone lineage of these derived cells will be followed by CRISPR barcoding or high-throughput imaging of fluorescent-based reporter cell expression. In addition, we will set up innovative tumor-on-chip models, allowing the reconstitution of the tumor microenvironment, including the tumor interface with the matrix, the blood vessels, the cancer-associated fibroblasts, and the immune cells. These innovative models will avoid the use of animal models, which are currently poorly accepted by society. DTPs obtained from these different models will be analyzed using high-throughput single-cell technologies, including transcriptomic and epigenomic, performed in the dedicated platforms of Lille University. Single-cell analyses are particularly suited for studying the small contingent of the cell population that has adapted to
treatment. This high-throughput data will be interpreted by original mathematical and bioinformatic methodologies allowing the modeling of regulatory networks. In parallel, we will develop original mathematical modeling of this biological process.
Functional involvement of the main regulatory pathways identified previously will be determined by genetic invalidation or pharmacological approaches with a particular focus on epigenetic, immunological regulations, stages of dedifferentiation into stem cells, cellular senescence, or modification of intracellular signal transduction pathways, all biological processes being studied by the biology teams gathered around the project.For these four bad prognosis cancers still treated by chemotherapies, our ambition is to identify, through this approach, new therapeutic targets that we will seek to inhibit, either by repositioning known active molecules or by designing new molecules, alone or in combination, to block the adaptation stages to treatment or subsequently eliminating drug-tolerant persister cells.